CRISPR:
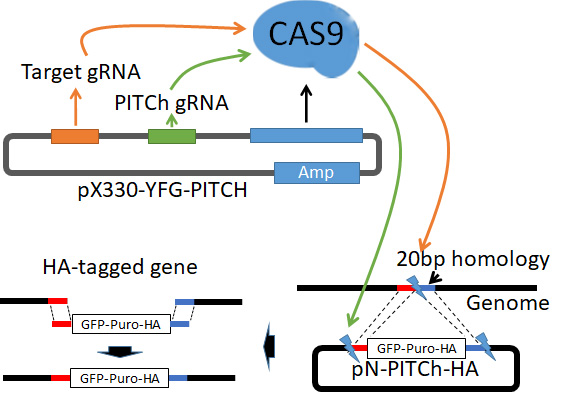
Work in yeast models has tremendously benefited from the insertion of epitope or fluorescence tags at the native gene locus to study protein function and behavior under physiological conditions. In contrast, work in mammalian cells largely relied on the overexpression of tagged proteins because high quality antibodies are only available for a fraction of the mammalian proteome. CRISPR/Cas9-mediated genome editing has recently emerged as a powerful genome-modifying tool, which can also be exploited to insert various tags and fluorophores at gene loci to study the physiological behavior of proteins in most organisms, including mammals. Here we describe a versatile tool set for rapid tagging of endogenous proteins. The strategy utilizes CRISPR/Cas9 and microhomology-mediated end-joining (MMEJ) repair for efficient tagging. We provide tools to insert 3xHA (hemagglutinin), 6xHisFLAG, HBTH, mCherry, GFP, and the AID tag for compound induced protein depletion. This approach and the developed tools should greatly facilitate functional analysis of proteins in their native environment.